How do immune cells spread?
When you picture a biological cell, you probably imagine some roughly spherical blob filled with all those strangely-named organelles you had to memorize in high school. But in reality, cells are dynamic objects, changing shape within seconds to perform different tasks. Immune cells in particular are capable of deforming rapidly, which they need to do for many of their main functions. Whenever you have some injury or infection, white blood cells respond to chemical cues as they travel through the blood stream, causing them to stick to the blood vessel wall, flatten out on the surface, and crawl out of the vessel (by squeezing quite dramatically!) into the surrounding tissue. They then crawl through the tissue and track down any foreign invaders (pathogens) such as bacteria, fungi, or viruses. Upon finding any of these pathogens, they will stick to the pathogenic surface and begin the process of phagocytosis (which literally means “cell eating”). You can see a few examples of phagocytosis by human neutrophils (the most abundant white blood cells) in the video below. This is previous work from Dr. Heinrich and colleagues, where they “fed” pathogenic particles to neutrophils and watched the process of consumption. In this video, the “pathogenic particles” are very small plastic beads coated with IgG antibodies.
Movie 1: Examples of human neutrophils consuming polystyrene microspheres coated with IgG (from Herant, Heinrich, and Dembo 2006)
From the brief story outlined above, you can hopefully appreciate that sticking to surfaces and spreading out in a controlled manner are crucial to the everyday life of human immune cells. How exactly do they achieve this, particularly the monumental feat of phagocytosis? The “sticking” part (referred to as “adhesion”) is a bit less mysterious; in the case of phagocytosis, receptors on the cell surface bind to molecules present on pathogen surfaces, providing a strong connection like glue. Many of the molecules these receptors bind to actually come from the bloodstream and label pathogen surfaces; a famous example of this which we focus on here is antibodies, mainly immunoglobin G (IgG). But once the cell sticks to a surface, how does it spread out and eventually consume the pathogen?
Figure 1: Phagocytosis as a “zippering” process. The front of the cell (white body) forms attachments to the pathogen (gray body) as it spreads. Figure from Swanson and Hoppe, 2004, Journal of Leukocyte Biology
There are two main hypotheses about how the cell spreads over a pathogen surface. In both conceptual models, linkages between receptors and the pathogen surface act like a sort of molecular “zipper”, as pictured in Figure 1.
Briefly, these models are as follows:
Brownian zipper: Spreading is powered solely by the energy gained from receptor binding. In this model, the cell is analogous to a water droplet spreading on a surface. No extra force is needed; the droplet (or the cell) will spread simply because it finds the surface to be sufficiently sticky.
Protrusive zipper: Spreading is driven by forces exerted by the cell itself. The main forces we think of in this context arise from the cell’s cytoskeleton pushing on the cell boundary. In particular a protein called “actin” forms filaments which can elongate and exert an outward pressure on the membrane, causing the cell to protrude.
Note that the protrusive zipper still requires adhesion to the pathogen surface to direct the cell around the pathogen; otherwise it would simply protrude in random directions rather than spread in the controlled manner you saw in the video above. The key distinction between these two zippers is what powers the actual cell deformation - adhesion or protrusion.
In the Heinrich Lab, we set out to answer this question using both computer simulations and experiments with human neutrophils. Our experiments were based on a simple principle: by changing the “stickiness” of the surface the cell is spreading on, we should be able to distinguish between the two models above. We did this by coating surfaces with different densities of IgG antibodies, ranging from tens of molecules per square micrometer to about 25,000 molecules per square micrometer (a micrometer, also called a micron, is 1/10,000th of a centimeter). If spreading were driven by adhesion only, as the Brownian zipper suggests, the stickiness of the surface should have a dramatic effect on both how fast and how far the cell spreads. But in the protrusive zipper model, this may not be the case; as long as the cell has enough molecules to stick to, one could imagine that it would spread quite efficiently, and further binding sites might not be of any help. As a rough analogy for this, imagine you’re on a rock climbing wall; once you have enough hand and footholds present, it won’t be of much help to add any more. Your energy as a rock climber, rather than the availability of holds, will be the limiting factor.
These experiments were conducted on flat surfaces coated with IgG, which acted as very, very large pathogens (effectively infinitely sized). In this case, the cell obviously can’t consume the entire surface, and the process of spreading is referred to as “frustrated phagocytosis” (yes, this is the technical term). In the first video above, the cell didn’t complete phagocytosis of the largest bead, so this is also an example of frustrated phagocytosis. The experimental setup is depicted in Figure 2 below.
Figure 2: Diagram of the frustrated phagocytosis experimental setup, modified from our experimental preprint (Francis et al 2022). The microscope setup is described in full in the paper; this setup allows for “reflection interference contrast microscopy”, which allows us to collect the high-contrast images of the cell “footprint” during spreading, as shown on the right hand side.
An example video of the spreading cells’ “footprints” (the contact region between the cell membrane and the coated glass coverslip) is also shown below. When using a particular imaging technique (reflection interference contrast microscopy), the contact regions show up as dark patches within the field of view.
Movie 2: This video shows the cell-substrate contact regions, or “footprints”, of neutrophils spreading on an IgG coated surface, imaged with reflection interference contrast microscopy. Movie from Francis et al 2022 preprint.
Figure 3: Measurements of spreading speeds and maximum contact areas during frustrated phagocytosis experiments. The spreading speed doesn’t change on higher IgG densities, and the maximum contact area only increases a small amount. Figure modified from Francis and Heinrich 2022.
Remarkably, our experiments showed that the neutrophils spread quite rapidly over any of the densities of IgG we tested. From our images of the spreading contact regions, we measured how fast the contact area (the area of the cell-surface contact region) grew over time. On all of our surfaces, the cells spread at about 3 square micrometers per second. This finding alone contradicts what we might expect from the Brownian zipper model.
We also quantified “how far” the cells spreading (what was the contact area when the cell decided to stop spreading), and did find a slight increase in maximum contact area on surfaces coated with higher densities of IgG. However, if spreading were powered by adhesion, the maximum contact area should increase dramatically as we increase IgG density. On surfaces coated with low densities of IgG, receptor binding alone shouldn’t provide enough force to allow the cell to deform considerably; therefore, we need some extra force to explain the spreading observed during phagocytosis, which we propose is provided by cell protrusion. The protrusive zipper model wins!
This is all well and good, but the above arguments remain qualitative and intuitive. To make this more rigorous and quantitative, we decided to simulate what spreading should look like based on the physics of the Brownian zipper model vs. the physics of the protrusive zipper model. Admittedly, this is no small task, but this sort of modeling was something I was passionate to explore in my time as a PhD student. Cells are incredibly complicated objects (recall all those weird organelles mentioned above), and even with state-of-the-art computing approaches, we can’t model an entire cell molecule by molecule. In this case, we simplified the cell structure considerably by treating it as a body of highly viscous fluid surrounded by an elastic boundary (“cell cortex”). There are a lot of interesting details about how we can choose to model biological cells, but I should save those for another blog post. For now, by rough analogy, it would suffice to imagine that we model the cell as something like a very small water balloon filled with a highly viscous fluid, like honey. The model is sketched out in Figure 4 below.
Figure 4: We model the spreading neutrophil as a body of highly viscous fluid with different stresses acting at the cell boundary. The diagram on the top right is essentially a free-body diagram, like you may have encountered when solving force balance problems in basic physics. In this case, the system is quite complicated overall, so we need to use a computer to compute a numerical solution, which gives us the fluid velocity and pressure, as plotted on the bottom right. Figure is modified from Francis and Heinrich 2022.
We started by modeling the Brownian zipper model, capturing the physical effects of adhesive attraction by assuming the cell membrane is attracted to the flat surface with a strength that depends on the overall density of IgG molecules. In a way, adhesion acts as a force which “pulls” the cell onto the surface. Not too surprisingly, the model cell spreads slowly and not too far for the lower densities of IgG, but if we increase IgG density, we observe rapid spreading to larger contact areas. This is exactly what we expect if adhesion itself drives the spreading, but it doesn’t at all match what we measured in our experiments.
Movie 3: Model predictions of how cells spread if spreading is driven by adhesion alone (Brownian zipper model). Here, the density of IgG is represented as a percentage in the title of the graph. Movie is from Francis and Heinrich 2022.
We next turned our attention to the protrusive zipper model, assuming that rather than being “pulled” onto the surface, the cell membrane is pushed outwards by active protrusive forces. Note that the cell still sticks to the surface, but adhesion does not act as an attractive force “pulling” the cell onto the surface. In this case, the cell spreads quite nicely, forming protrusions which look similar to the videos from real cells shown above. Time to pack up and call it a day, right?
Movie 4: Model predictions of how cells spread if spreading is driven by protrusion alone (protrusive zipper model). Here, we assume the cell can attach to the surface anywhere it wants, and the IgG density does not govern the forces acting on the spreading cell. Movie is from Francis and Heinrich 2022.
Not quite. The above simulation was conducted assuming that the cell can attach to the surface anywhere it wants to, which should strike you as somewhat suspicious. If there’s enough IgG on the surface, perhaps we could get away with this, but for the lower densities we tested, the assumption is utterly unrealistic. Return to the analogy of the rock-climber. If there are enough hand and foot holds available everywhere on the wall, you can safely assume that a hold will be readily available wherever you need it. But if those hand and foot holds are spaced out, you absolutely need to know the exact location of the next hold; this will be key to determining how far you’ll be able to make it up the wall!
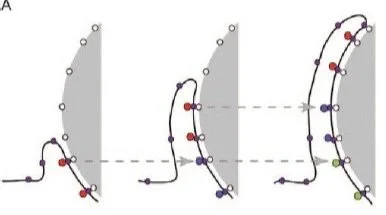
With that in mind, we can modify the implementation of the protrusive zipper model, accounting for the discrete nature of adhesion. After all, we still want to test for any effects of changing the density of binding sites on the surface, and we do know that there was an effect on the maximum contact area in our experiments. You can see the revised model depicted below. Rather than saying that the cell can stick anywhere it wants to on the surface, we say that it can only form new attachments in specified locations (depicted as dots on the surface in the drawing below).
Figure 5: A zoomed-in depiction of a neutrophil spreading over discrete binding sites. The cell can spread further on a surface coated with higher densities of IgG, just as you would likely be able to climb higher on a rock wall with plenty of hand and foot holds available.
We also assumed that the protrusive force exerted by the cell decreases when the cell goes longer between forming one attachment and grabbing onto the next one. The rationale here is that the cell requires biochemical stimulus from adhesion to trigger protrusion. The exact details behind that involve complex cell signaling processes which we don’t model, so in this case our assumption is just meant to capture this qualitative phenomenon.
The video below is a zoomed-in depiction of the model cell spreading over discrete binding sites (this is my favorite video from the paper!). As you can hopefully see, when the binding sites are more spaced out, the cell spreads efficiently initially, but at some point, it can’t quite reach that next binding site and it “gives up”. As you might expect, when we put the binding sites closer together, the cell can spread further. This matches quite well with what we see in our experiments; cells spread at the same speed regardless of ligand density, but the maximum contact area is higher when the IgG density is higher.
Movie 5: Model predictions of how cells spread if spreading is driven by protrusion alone (protrusive zipper model), with the added consideration of discrete adhesion sites. The IgG density is depicted graphically by the density of dots on the surface, and also is specified in the title of the plot. Movie is from Francis and Heinrich 2022.
If I’ve done my job here, this should give you an idea of how applying physical models to cells can give us real insights on how they work. In our case, by comparing the outcomes of experiments and computational models, we concluded that protrusion, not adhesion, is what powers immune cell spreading during phagocytosis. Additionally, simple physical considerations such as the spacing between IgG molecules allow us to explain what we see in our experiments. To me, this is a good example of how simulations and experiments can work together to inform our understanding of biology. This is an overarching goal I have in my scientific research career - to interface between simulations/theory and hands-on experiments to really understand how cells work at a basic, physical level. I’ll be excited to share more examples of this in the future, thanks for reading!
Our recent studies discussed in this post:
Computational modeling paper in PLOS Computational Biology: Francis EA, Heinrich V (2022) Integrative experimental/computational approach establishes active cellular protrusion as the primary driving force of phagocytic spreading by immune cells. PLoS Comput Biol 18(8): e1009937. https://doi.org/10.1371/journal.pcbi.1009937
Experimental preprint manuscript: Francis, E., Xiao, H., Teng, L. H. & Heinrich, V. Mechanisms of frustrated phagocytic spreading of human neutrophils on antibody-coated surfaces. bioRxiv 2022.02.18.481104 (2022) doi:10.1101/2022.02.18.481104.
Other references:
Herant, M., Heinrich, V. & Dembo, M. Mechanics of neutrophil phagocytosis: experiments and quantitative models. J Cell Sci 119, 1903–1913 (2006).
Swanson, Joel A.; Hoppe, Adam D. (2004). "The coordination of signaling during Fc receptor‐mediated phagocytosis." Journal of Leukocyte Biology 76(6): 1093-1103.
Heinrich, V. Controlled One-on-One Encounters between Immune Cells and Microbes Reveal Mechanisms of Phagocytosis. Biophys J 109, 469–476 (2015). [introduces and discusses the Brownian zipper vs. the protrusive zipper]